 |
| In Cycle Sequencing using a DNA auto sequencer, basecalling was started
at25-30 bases from the 3'end of the sequencing primer. On the other hand,
in Transcriptional Sequencing, basecalling was started at 50 bases from
the initiation site. Therefore there are some uncertain areas in the 5'
end sequences using the current plasmid DNA with promoter sequences in
Transcriptional Sequencing. To make up for its defect, we developed the
cloning vector "pTS1" DNA, which is the optimal solution for
accurate Transcriptional Sequencing. The cloning vector "pTS1" for Transcriptional Sequencing is derived from pUC19 and you can prepare your target DNA with the T3, T7 promoter for Transcriptional Sequencing. pTS1 includes lacZ'ƒ¿ as the part of ß-galactositase in E.coli. The lacZ'ƒ¿ fragment includes multiple cloning sites (MCS). If you insert your target DNA into MCS, you can distinguish colonies in the blue/white selection. [The advantages of pTS1]
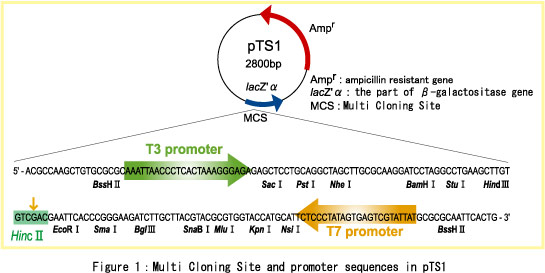 It can also be used for Cycle Sequencing using some sequencing primers listed in Figure 2. When you have cloning steps with pTS1, you can choose either Cycle Sequencing or Transcriptional Sequencing depending on whichever method you prefer. 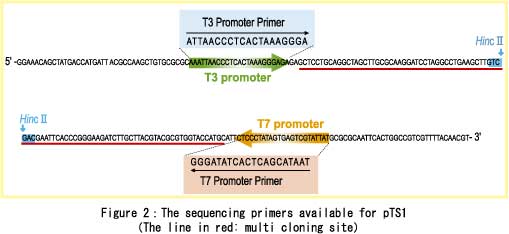 [The host that we recommend for pTS1] We recomemend E.coli JM109 to use pTS1. As for sales in other countries, please wait for this product to reach you in the near future... For more information about our product,
contact us:
|